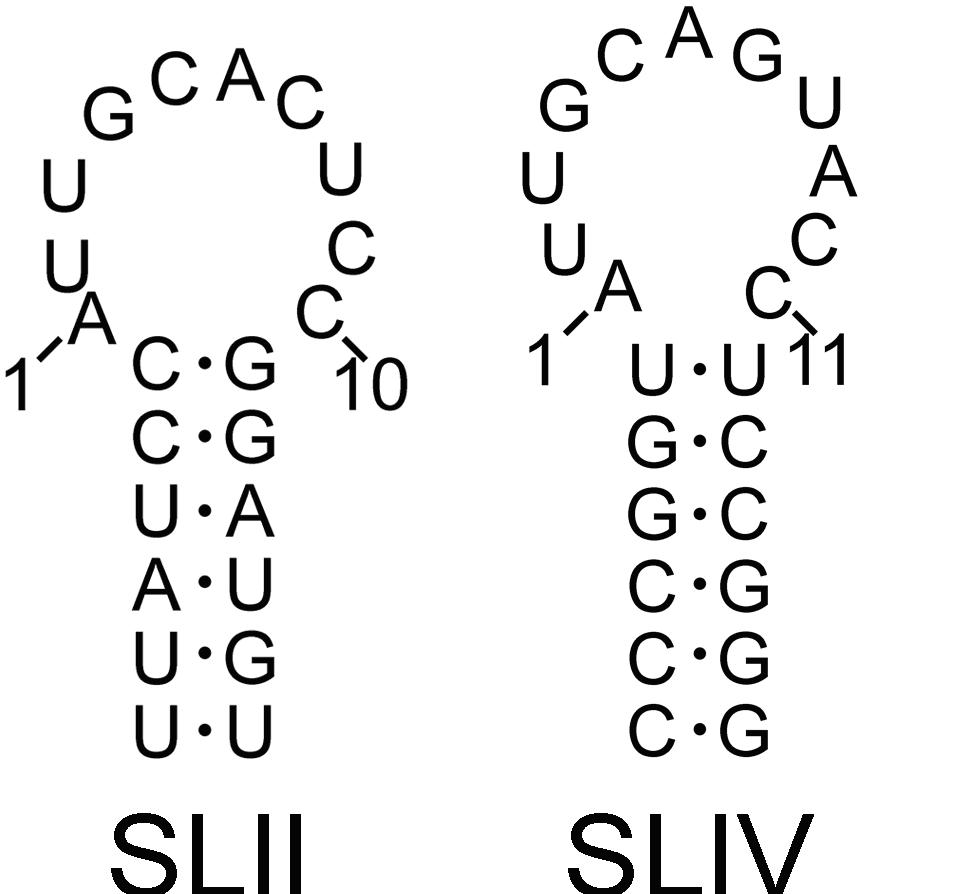Lab Projects
RNA-Binding Protein Allostery
SNF is an RNA binding protein from Drosophila that is found in both the U1 and U2 snRNPs. It uses the common eukaryotic RNA Recognition Motif (RRM) to bind RNA stemloops in U1 and U2 snRNA, and is a member of the larger U1A/U2B”/SNF family. We have constructed a phylogenetic family tree of these proteins as far back as the urbilaterian divergence, which shows that SNF’s branch is distinct from that of the vertebrate U1A/U2B” proteins. The evolution of this family in vertebrates resulted in two proteins: U1A that binds to U1 snRNA, and U2B” that binds to U1 and U2 snRNA (in vivo, it’s found only in the U2 snRNP). In most other eukaryotic organisms, there is a single protein, exemplified by SNF.
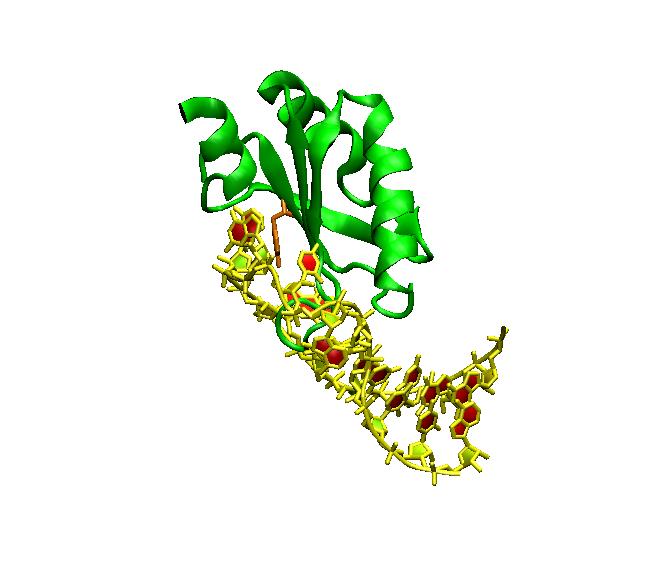
Human SLII and SLIV. Drosophila SLIV has a U:G loop closing base pair. A cocrystal of the complex of U2B” RRM (green) bound to SLIV RNA (yellow) (pdb 1A9N). Note the green loop of U1A that is inserted in the RNA loop. To date, there is no comparable co-crystal of SNF.
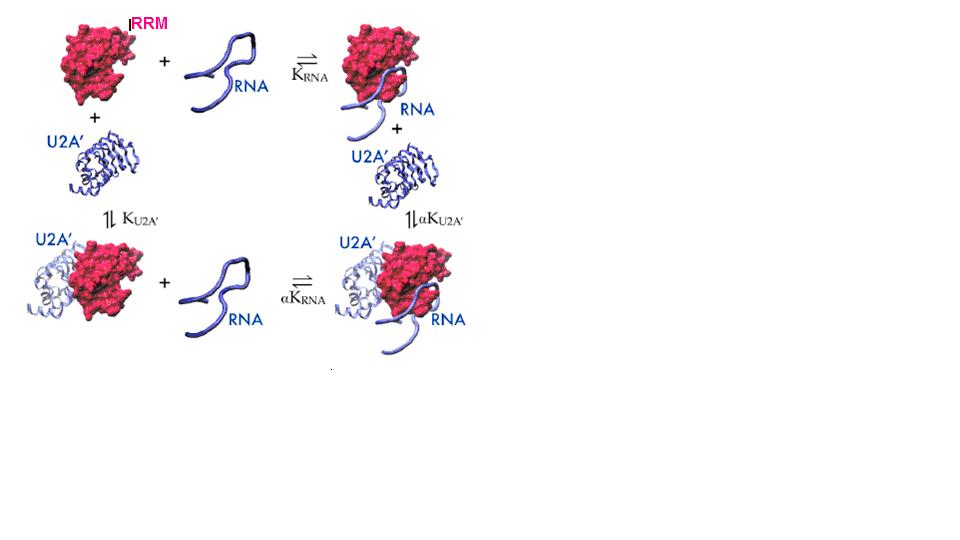
SNF binds to both U1 snRNA Stemloop II (SLII) and U2 snRNA Stemloop IV (SLIV), but binding affinity is greater for the SNF:SLII complex. In the context of the nucleus, where the U1 snRNP is present in large excess over the U2 snRNP, it might be expected that most of the SNF protein would be sequestered in the U1 snRNP. However, SNF also binds to the U2A’ protein, and the affinity of the SNF:U2A’ complex for SLIV is greatly enhanced over affinity for SLII, thus ensuring that SNF is found in both snRNPs. A thermodynamic cycle of the binary/ternary complexes is shown below, and is the basis of our current investigations that relate binding affinity and to allosteric regulation. Here, we can define SNF as the macromolecule that binds two ligands: RNA and U2A’, and we are using NMR, ITC, and computational methods to uncover the molecular mechanism that underlies this change in SNF RNA binding preferences.
Varkud Satellite Virus Stemloop V
This five nucleotide RNA loop forms a modified U-turn that is stabilized in the presence of Mg2+ ions. It is one element of the ~160 nucleotide ribozyme that is responsible for processing the entire RNA strand for packaging. The ribozyme can be excised and studied independently, and stemloop V has been described by NMR (Campbell & Legault, 2005). SL V interacts with SL I to form a kissing hairpin complex that positions the ribozyme active site with respect to the cleavage site.
The uUGACUa hairpin adopts a U-turn conformation with the first four nucleotides of the loop (UGAC) forming the core of the structure. The fifth U nucleotide is extruded from the loop, a conformation that is stabilized by Mg2+ ions. We are using this stemloop as a model system for RNA dynamics measurements by NMR and 2-aminopurine fluorescence and computational methods. We find that without Mg2+, the RNA samples conformational space, and accesses structures that include the Mg2+-bound state. While there is stacking of the GAC nucleobases, the stacking is not stable; without Mg2+, the purine bases spend at least 60% of their time unstacked, but with Mg2+, the A is predominantly stacked while the G is predominantly unstacked. Computational experiments using AMBER capture some of this behavior, but overstacks the G and A.
This example of a U-turn RNA hairpin loop offers an opportunity to perturb its sequence, specifically the fourth nucleobase, to measure the effect on its stability and structure, and Mg2+ dependence. Such information will be useful to anticipate the structure and structural stability of this class of U-turns, and leads to the construction of other classes. We think there should be four classes: the triloops with the characteristic UNR sequence (U is uridine, N anything, R purine); tetraloops with UNRX sequences; and loops with sequences UNRXn, where n>1. We predict that triloops will require no divalent ions for structure/stability, while all UNRXn loops will be floppy without Mg2+ but will adopt a U-turn with Mg2+ by extruding the 3′ nucleotides.
GTPase Center RNA
This 60 nucleotide RNA has been studied by dissection into its stemloop elements, and also intact. A crystal structure of the E coli form shows an intricate 3D fold that includes loop-loop interactions and one ‘chelated’ K+ ion.
More Coming Soon!